Anti-Cancer Effects of Glaucarubinone in the Hepatocellular Carcinoma Cell Line Huh7 via Regulation of the Epithelial-To-Mesenchymal Transition-Associated Transcription Factor Twist1
Abstract
:1. Introduction
2. Results
2.1. GCB Showed Low Cytotoxicity
2.2. Malignant Traits of Huh7 Cells Were Suppressed by GCB
2.3. GCB Altered MMP Activities
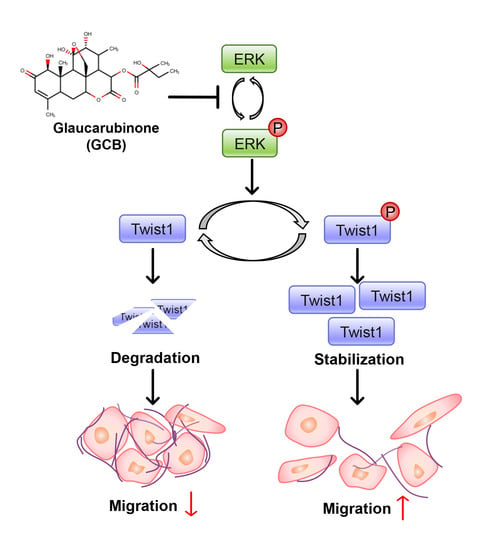
2.4. Twist1 and ERK Activities Were Inhibited by GCB
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Wound Healing Screening
4.3. Antibodies
4.4. GCB Isolation and Preparation
4.5. Cell Viability Assay
4.6. Wound Healing Assay
4.7. Transwell Migration and Invasion Assay
4.8. Soft Agar Colony Formation Assay
4.9. Gelatin Zymography
4.10. MMP Activity Assay
4.11. Three-Dimensional Spheroid Invasion Assay
4.12. Transfection
4.13. Twist1 siRNA Transfection
4.14. Immunoblotting Analysis
4.15. qRT-PCR Analysis
4.16. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GCB | Glaucarubinone |
| HCC | Hepatocellular carcinoma |
| ERK | Extracellular signal-regulated kinase |
| JNK | c-Jun N-terminal kinase |
| EMT | Epithelial-to-mesenchymal transition |
| MMP | Matrix metalloproteinase |
| MAPK | Mitogen-activated protein kinase |
| PAK | p21-activated kinase |
References
- Bosch, F.X.; Ribes, J.; Díaz, M.; Cléries, R. Primary liver cancer: Worldwide incidence and trends. Gastroenterology 2004, 127, S5–S16. [Google Scholar] [CrossRef]
- Suhail, Y.; Cain, M.P.; Vanaja, K.; Kurywchak, P.A.; Levchenko, A.; Kalluri, R.; Kshitiz. Systems Biology of Cancer Metastasis. Cell Syst. 2019, 9, 109–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szmulewitz, R.; Taylor, J.; Rinker-Schaffer, C. Metastatic Colonization. In Encyclopedia of Cancer; Springer: Berlin/Heidelberg, Germany, 2015; pp. 2797–2799. [Google Scholar]
- Frixen, U.H.; Behrens, J.; Sachs, M.; Eberle, G.; Voss, B.; Warda, A.; Lochner, D.; Birchmeier, W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 1991, 113, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Berx, G.; Cleton-Jansen, A.M.; Nollet, F.; De Leeuw, W.J.F.; Van De Vijver, M.J.; Cornelisse, C.; Van Roy, F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995, 14, 6107–6115. [Google Scholar] [CrossRef]
- Liu, Y.N.; Lee, W.W.; Wang, C.Y.; Chao, T.H.; Chen, Y.; Ji, H.C. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 2005, 24, 8277–8290. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.; Restuccia, D.F.; Lan, Q.; Hynx, D.; Dirnhofer, S.; Hess, D.; Rüegg, C.; Hemmings, B.A. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-β signaling axes. Cancer Discov. 2012, 2, 248–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivertsen, S.; Hadar, R.; Elloul, S.; Vintman, L.; Bedrossian, C.; Reich, R.; Davidson, B. Expression of Snail, Slug and Sip1 in malignant mesothelioma effusions is associated with matrix metalloproteinase, but not with cadherin expression. Lung Cancer 2006, 54, 309–317. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Zhou, J.; Fu, J.; He, T.; Qin, J.; Wang, L.; Liao, L.; Xu, J. Phosphorylation of serine 68 of twist1 by MAPKs stabilizes twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 2011, 71, 3980–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beutler, J.A.; Kang, M.I.; Robert, F.; Clement, J.A.; Pelletier, J.; Colburn, N.H.; McKee, T.C.; Goncharova, E.; McMahon, J.B.; Henrich, C.J. Quassinoid inhibition of AP-1 function does not correlate with cytotoxicity or protein synthesis inhibition. J. Nat. Prod. 2009, 72, 503–506. [Google Scholar] [CrossRef] [Green Version]
- Huynh, N.; Beutler, J.A.; Shulkes, A.; Baldwin, G.S.; He, H. Glaucarubinone inhibits colorectal cancer growth by suppression of hypoxia-inducible factor 1α and β-catenin via a p-21 activated kinase 1-dependent pathway. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Yeo, D.; Huynh, N.; Beutler, J.A.; Christophi, C.; Shulkes, A.; Baldwin, G.S.; Nikfarjam, M.; He, H. Glaucarubinone and gemcitabine synergistically reduce pancreatic cancer growth via down-regulation of P21-activated kinases. Cancer Lett. 2014, 346, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Matsushima, H.; Nishibu, A.; Clausen, B.E.; Takashima, A. Dual Therapeutic Efficacy of Vinblastine as a Unique Chemotherapeutic Agent Capable of Inducing Dendritic Cell Maturation. Cancer Res. 2009, 69, 6987–6994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Liu, S.; Che, X.; Hou, K.; Ma, Y.; Li, C.; Wen, T.; Fan, Y.; Hu, X.; Liu, Y.; et al. Bufalin inhibits TGF-β-induced epithelial-to-mesenchymal transition and migration in human lung cancer A549 cells by downregulating TGF-β receptors. Int. J. Mol. Med. 2015, 36, 645–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, M.-W.; Chen, C.-H.; Chang, Y.-L.; Teng, C.-M.; Pan, S.-L. α-Tomatine-Mediated Anti-Cancer Activity In Vitro and In Vivo through Cell Cycle- and Caspase-Independent Pathways. PLoS ONE 2012, 7, e44093. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Gong, Z.; Wu, Q.; Su, Q.; Yang, T.; Yu, R.; Xu, R.; Zhang, Y. Homoharringtonine suppresses tumor proliferation and migration by regulating EphB4-mediated β-catenin loss in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 632. [Google Scholar] [CrossRef]
- Chen, J.; Jiao, D.; Li, Y.; Jiang, C.; Tang, X.; Song, J.; Chen, Q. Mogroside V Inhibits Hyperglycemia-induced Lung Cancer Cells Metastasis through Reversing EMT and Damaging Cytoskeleton. Curr. Cancer Drug Targets 2019, 19, 885–895. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef]
- Suarez-Carmona, M.; Lesage, J.; Cataldo, D.; Gilles, C. EMT and inflammation: Inseparable actors of cancer progression. Mol. Oncol. 2017, 11, 805–823. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Chang, J.T.; Andrechek, E.R.; Matsumura, N.; Baba, T.; Yao, G.; Kim, J.W.; Gatza, M.; Murphy, S.; Nevins, J.R. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene 2009, 28, 2796–2805. [Google Scholar] [CrossRef] [Green Version]
- Scheau, C.; Badarau, I.A.; Costache, R.; Caruntu, C.; Mihai, G.L.; Didilescu, A.C.; Constantin, C.; Neagu, M. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal. Cell. Pathol. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol. Rep. 2009, 21, 1323–1333. [Google Scholar] [CrossRef]
- Rajesh, E.; Sankari, L.; Malathi, L.; Krupaa, J. Naturally occurring products in cancer therapy. J. Pharm. Bioallied Sci. 2015, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.-S.S.; Gao, W.; Chan, J.Y.-W.W. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Hoti, S.L.; Nazeer, Y.; Hegde, H.V. Glaucarubinone sensitizes KB cells to paclitaxel by inhibiting ABC transporters via ROS-dependent and p53-mediated activation of apoptotic signaling pathways. Oncotarget 2016, 7, 42353–42373. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Nakagawa-Goto, K.; Lang, J.Y.; Kim, Y.; Lai, C.Y.; Goto, M.; Sakurai, N.; Taniguchi, M.; Akiyama, T.; Morris-Natschke, S.L.; et al. Antitumor agents. 282. 2′-(R)-O-acetylglaucarubinone, a quassinoid from Odyendyea gabonensis as a potential anti-breast and anti-ovarian cancer agent. J. Nat. Prod. 2010, 73, 1553–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, D.; Huynh, N.; Beutler, J.A.; Baldwin, G.S.; He, H.; Nikfarjam, M. Glaucarubinone Combined with Gemcitabine Improves Pancreatic Cancer Survival in an Immunocompetent Orthotopic Murine Model. J. Investig. Surg. 2016, 29, 366–372. [Google Scholar] [CrossRef]
- López-Camarillo, C.; Ocampo, E.A.; Casamichana, M.L.; Pérez-Plasencia, C.; Álvarez-Sánchez, E.; Marchat, L.A. Protein kinases and transcription factors activation in response to UV-radiation of skin: Implications for carcinogenesis. Int. J. Mol. Sci. 2012, 13, 142–172. [Google Scholar] [CrossRef]
- Cho, K.H.; Jeong, K.J.; Shin, S.C.; Kang, J.; Park, C.G.; Lee, H.Y. STAT3 mediates TGF-β1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett. 2013, 336, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Besser, A.H.; Wander, S.A.; Sun, J.; Zhou, W.; Wang, B.; Ince, T.; Durante, M.A.; Guo, W.; Mills, G.; et al. Cytoplasmic p27 promotes epithelial-mesenchymal transition and tumor metastasis via STAT3-mediated Twist1 upregulation. Oncogene 2015, 34, 5447–5459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Ren, Y.; Jia, X.; Liang, P.; Lou, W.; He, L.; Li, M.; Sun, S.; Wang, H. Twist overexpression promoted epithelial-to-mesenchymal transition of human peritoneal mesothelial cells under high glucose. Nephrol. Dial. Transplant. 2012, 27, 4119–4124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Hu, H.G.; Huang, J.; Zou, Q.; Wang, J.; Liu, M.Q.; Zhao, Y.; Li, G.Z.; Xue, S.; Wu, Z.S. Expression and correlation of Twist and gelatinases in breast cancer. Exp. Ther. Med. 2013, 6, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.H.; Yang, X.Q.; Wang, B.C.; Liu, S.P.; Wang, F.B. Overexpression of twist and matrix metalloproteinase-9 with metastasis and prognosis in gastric cancer. Asian Pac. J. Cancer Prev. 2013, 14, 5055–5060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterman, P.G.; Ampofo, S.A. Cytotoxic quassinoids from Odyendyea gabonensis stem bark: Isolation and high-field NMR. Planta Med. 1984, 50, 261–263. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, J.; Ha, J.; Kang, E.; Yoon, H.; Lee, S.; Ryu, S.Y.; Kim, K.; Cho, S. Anti-Cancer Effects of Glaucarubinone in the Hepatocellular Carcinoma Cell Line Huh7 via Regulation of the Epithelial-To-Mesenchymal Transition-Associated Transcription Factor Twist1. Int. J. Mol. Sci. 2021, 22, 1700. https://doi.org/10.3390/ijms22041700
Seo J, Ha J, Kang E, Yoon H, Lee S, Ryu SY, Kim K, Cho S. Anti-Cancer Effects of Glaucarubinone in the Hepatocellular Carcinoma Cell Line Huh7 via Regulation of the Epithelial-To-Mesenchymal Transition-Associated Transcription Factor Twist1. International Journal of Molecular Sciences. 2021; 22(4):1700. https://doi.org/10.3390/ijms22041700
Chicago/Turabian StyleSeo, Jihye, Jain Ha, Eunjeong Kang, Haelim Yoon, Sewoong Lee, Shi Yong Ryu, Kwonseop Kim, and Sayeon Cho. 2021. "Anti-Cancer Effects of Glaucarubinone in the Hepatocellular Carcinoma Cell Line Huh7 via Regulation of the Epithelial-To-Mesenchymal Transition-Associated Transcription Factor Twist1" International Journal of Molecular Sciences 22, no. 4: 1700. https://doi.org/10.3390/ijms22041700